Karin List
Office address
540 East Canfield, Room 6332
Detroit, Michigan 48201
Office phone
313-577-1034
Biography
Extracellular proteases can degrade extracellular matrix proteins and reshape the tissue microenvironment as well as cleave and activate signaling molecules such as growth factors and their receptors. These processes are essential for normal physiological tissue development, remodeling, and repair. On the flip side, tissue malfunction and tissue destruction due to dysregulated extracellular proteolysis characterize many pathological conditions including cancer. Extracellular proteolytic processes are critically involved in tumor growth, invasion, and dissemination of cancer cells to other organs.
Proteolysis in the extracellular/pericellular environment is mediated by about 300 different proteases in humans, of which approximately one-third are directly anchored to the plasma membrane. We are particularly interested in a family of cell-surface-anchored proteases: the type II transmembrane serine proteases (TTSPs) and their role in tissue remodeling during epithelial development and carcinogenesis.
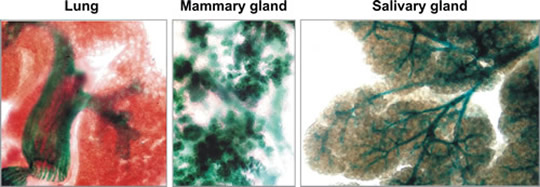 |
Epithelial expression of the type II transmembrane serine protease matriptase.
A knock-in mouse with a promoterless β-galactosidase marker gene inserted into the matriptase locus is used as a unique tool for assessing endogenous matriptase expression in various tissues. Matriptase (cyan/green) is expressed in the epithelium of lung, mammary gland, and salivary gland as determined by X-gal staining of mouse organ whole mounts.
Understanding the physiological role of extracellular proteases in tissue development and homeostasis is important in order to pinpoint how dysregulated proteolysis can cause or contribute to cancer progression. The motivation behind parallel investigations of normal physiology and pathology is the idea that carcinogenesis often involves pathways, including proteolytic pathways, that are important in normal development and have gone awry in cancer. Generation and characterization of mouse models, including models of human cancer, play an integral role in our research. We use knock-out and transgenic mice for selected extracellular proteases and protease inhibitors as unique tools to identify critical proteolytic pathways in health and disease.
 |
Current Lab Personnel:
Carly Martin, PhD Candidate
Kimberley Sala-Hambrick, Research Assistant
Jacob Mackinder, Research Assistant
Hiwot Lessanework, Undergraduate Student, IMSD Program
Training
The National Institutes of Health, NIDCR, Bethesda, Maryland, USA (laboratory of Dr. Thomas H. Bugge)
Education
M. Sc., University of Copenhagen, Denmark, 1996 (cell biology)
Ph.D., University of Copenhagen, Denmark, 2000 (genetics, biochemistry)
Publications
- Tanabe LM, List K. "The Role of Type II Transmembrane Serine Protease Mediated Signaling in Cancer” (2016), FEBS Journal, Nov 21. doi: 10.1111/febs.13971. [Epub ahead of print].
- Zoratti GL, Tanabe LM, Hyland TE, Duhaime MG, Colombo E, Leduc R, Marsault E, Johnson MD, Lin CY, Boerner J, Lang JE, List K. Matriptase regulates c-Met mediated proliferation and invasion in inflammatory breast cancer (2016), Oncotarget. 6;7(36):58162-58173.
- Murray AS, Varela FA, and List K. Type II transmembrane serine proteases as potential targets for cancer therapy (2016). Biol Chem, 397(9):815-26.
- Duhaime MJ, Murray AS, Varela FA, Zoratti GL and List K. Cell surface Human Airway Trypsin- like Protease is lost during squamous cell carcinogenesis (2016). J Cell Physiol, 231(7):1476-83
- Zoratti GL, Tanabe LM, Bergum C, Colombo C, Lang J, Molinolo A, Leduc R, Marsault E, Boerner J, List K. Targeting matriptase in breast cancer abrogates tumor progression via impairment of stromal-epithelial growth factor signaling (2015). Nature Communications, (15);6:6776.
Search PubMed for publications from the List Lab
